Romana Ďuráčiová, a student at FIT BUT, received two prestigious awards for the development and presentation of the Foldify graphical interface at the Excel@FIT 2025 student conference. Kristián Kováč presented there the Genomic Data Infrastructure (GDI) project, focused on the secure processing and analysis of sensitive genomic data in an international collaboration environment. Both projects are being developed at the CERIT-SC Center under the leadership of Lukáš Hejtmánek.
 We are pleased to announce that Romana Ďuráčiová, a student at the Faculty of Information Technology, Brno University of Technology (FIT BUT), received two prestigious awards for her development and presentation of the Foldify graphical environment at the Excel@FIT 2025 student conference. The tool, developed at CERIT-SC under the supervision of Lukáš Hejtmánek, was awarded by both the expert jury and industry partners.
We are pleased to announce that Romana Ďuráčiová, a student at the Faculty of Information Technology, Brno University of Technology (FIT BUT), received two prestigious awards for her development and presentation of the Foldify graphical environment at the Excel@FIT 2025 student conference. The tool, developed at CERIT-SC under the supervision of Lukáš Hejtmánek, was awarded by both the expert jury and industry partners.
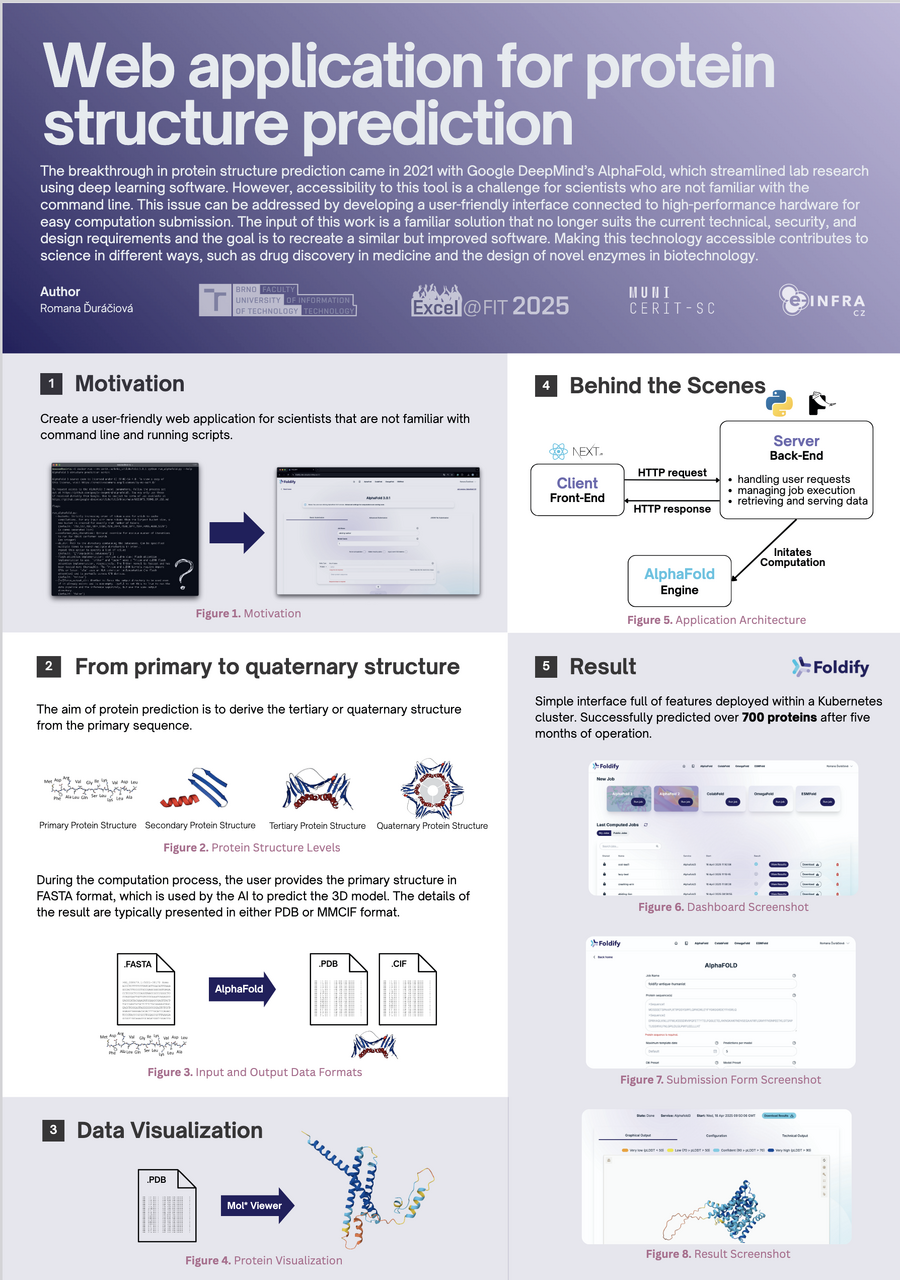
Foldify is an intuitive graphical environment designed for use with AlphaFold, a revolutionary technology for predicting protein 3D structures. AlphaFold, developed by DeepMind, made a major breakthrough in 2020 with the release of AlphaFold2, which achieved unprecedented accuracy in protein structure prediction—comparable to experimental methods like X-ray crystallography. This advancement was hailed as one of the greatest leaps in biology and contributed to the awarding of the Nobel Prize in Chemistry to the team involved in its development.
Building on AlphaFold2, AlphaFold3 was introduced in 2024 and expands prediction capabilities to full biomolecular complexes. In addition to proteins, it can model interactions with DNA, RNA, ligands, and ions—opening new avenues for drug discovery, understanding cellular regulatory mechanisms, and designing synthetic biomolecules. AlphaFold3 is a result of collaboration between DeepMind and Isomorphic Labs and represents a new benchmark in structural biology.
The motivation behind Foldify was to make the revolutionary AlphaFold technology accessible to a broader scientific community. Using AlphaFold can be challenging for many biologists due to the need for command-line skills and access to high-performance computing infrastructure. Foldify addresses this issue by offering a user-friendly graphical interface that enables easy task submission and visualization of results without technical knowledge, while connecting users to the powerful computing cluster operated by CERIT-SC.
Foldify runs in a Kubernetes environment managed by CERIT-SC and is designed to simplify access to protein structure prediction. It supports easy job submission, feedback, visualization, and downloading of predicted proteins. It includes optimized versions of AlphaFold, ColabFold, OmegaFold, and ESMFold. In just five months of operation, over 700 predictions have been successfully completed.
-
Web interface: https://foldify.cloud.e-infra.cz/
-
User guide: https://docs.cerit.io/en/docs/web-apps/foldify
 In addition to Romana, another FIT BUT student, Kristián Kováč, also participated in the conference, presenting the Genomic Data Infrastructure (GDI) project, which focuses on the secure processing and analysis of sensitive genomic data in the context of international collaboration. Although he did not receive an award, his contribution to our team is highly significant.
In addition to Romana, another FIT BUT student, Kristián Kováč, also participated in the conference, presenting the Genomic Data Infrastructure (GDI) project, which focuses on the secure processing and analysis of sensitive genomic data in the context of international collaboration. Although he did not receive an award, his contribution to our team is highly significant.
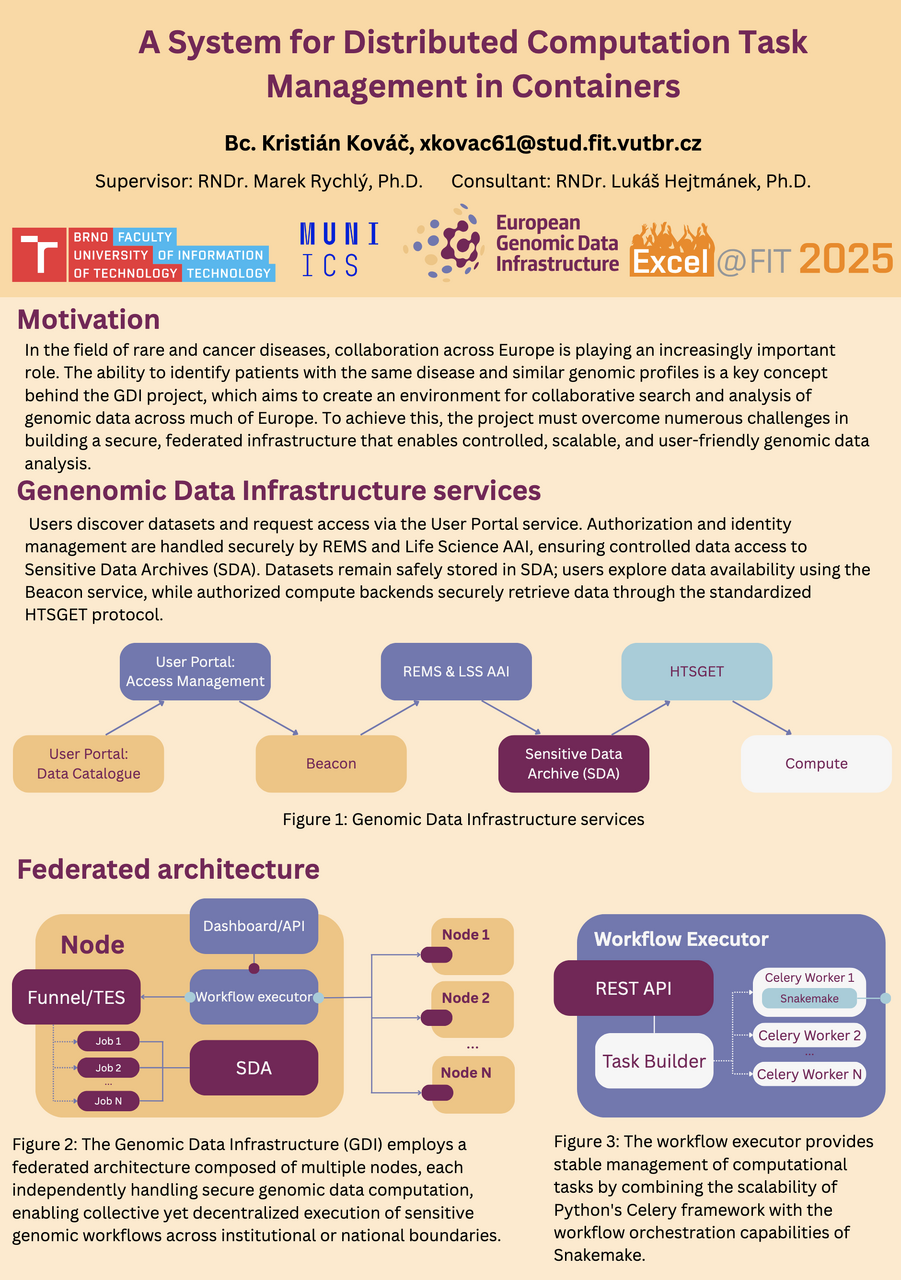
Under the supervision of Lukáš Hejtmánek from CERIT-SC, Kristián is developing a comprehensive portal and infrastructure to support secure workflows, including components for data submission, access rights management, and analysis.
The infrastructure adopts modern approaches such as OAuth2, GA4GH Visas, and employs tools like Snakemake, Kubernetes, and Celery for scalable and robust workflow management. The result is a federated platform that enables genomic data analysis across institutions while maintaining high levels of security and access control.
The system architecture is decentralized—individual compute nodes operate independently and store sensitive data locally in secure SDA repositories. Each node includes a Workflow Executor that manages the execution of computational tasks. Data processing occurs exclusively within authorized services, allowing workflows to run across institutions without compromising data. Every job execution is authenticated via token, enabling transparent work with sensitive data. Overall, the system offers a robust, scalable, and secure environment for collaborative genomic computing.
The GDI project thus makes a significant contribution to the goals of international initiatives such as B1MG (Beyond 1 Million Genomes) and 1+MG, which aim to interconnect genomic data across Europe for research and personalized medicine.
-
More about the project: https://gdi.onemilliongenomes.eu/

